CIQUIBIC-CONICET-UNC
Contact : +54 351 5353855
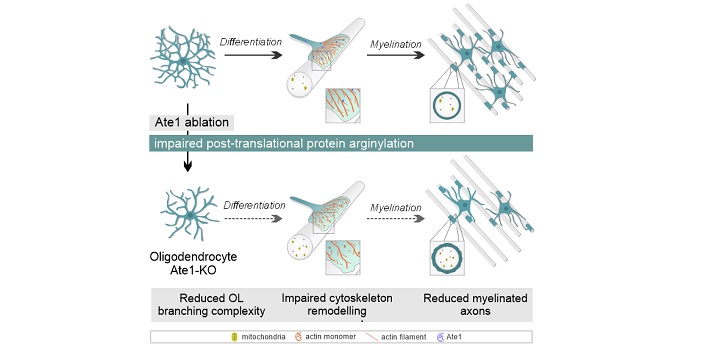
Addition of arginine (Arg) from tRNA can cause major alterations of structure and function of protein substrates. This post-translational modification, termed protein arginylation, is mediated by the enzyme arginyl-tRNA-protein transferase 1 (Ate1). Arginylation plays essential roles in a variety of cellular processes, including cell migration, apoptosis, and cytoskeletal organization. Ate1 is associated with neuronal functions such as neurogenesis and neurite growth. However, the role of Ate1 in glial development, including oligodendrocyte (OL) differentiation and myelination processes in the central nervous system, is poorly understood. The present study revealed a peak in Ate1 protein expression during myelination process in primary cultured OLs. Post-transcriptional downregulation of Ate1 reduced the number of OL processes, and branching complexity, in vitro. We conditionally ablated Ate1 from OLs in mice using 2′,3′-cyclic nucleotide 3′-phosphodiesterase-Cre promoter (“Ate1-KO” mice), to assess the role of Ate1 in OL function and axonal myelination in vivo. Immunostaining for OL differentiation markers revealed a notable reduction of mature OLs in corpus callosum of 14-day-old Ate1-KO, but no changes in spinal cord, in comparison with wild-type controls. Local proliferation of OL precursor cells was elevated in corpus callosum of 21-day-old Ate1-KO, but was unchanged in spinal cord. Five-month-old Ate1-KO displayed reductions of mature OL number and myelin thickness, with alterations of motor behaviors. Our findings, taken together, demonstrate that Ate1 helps maintain proper OL differentiation and myelination in corpus callosum in vivo, and that protein arginylation plays an essential role in developmental myelination.
Authors:
Article:
Palandri A., Bonnet L.V., Farias M.G., Hallak M.E:, Galiano M.R.. Glia. October 2021. https://doi.org/10.1002/glia.24107
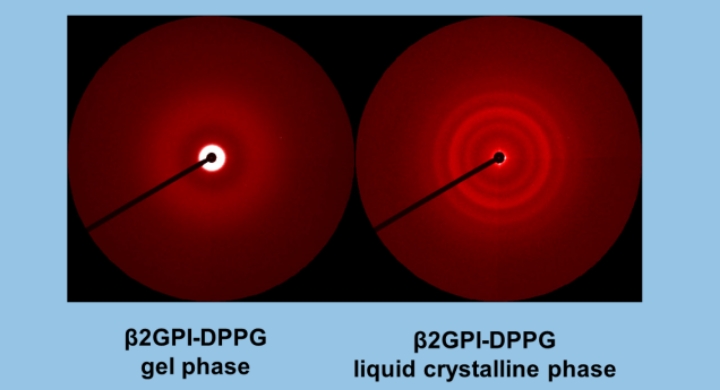
Rafael G. Oliveira, Mariana Paolorossi, Leide Passos Cavalcanti, Antonio Malfatti-Gasperini, Guillermo G. Montich

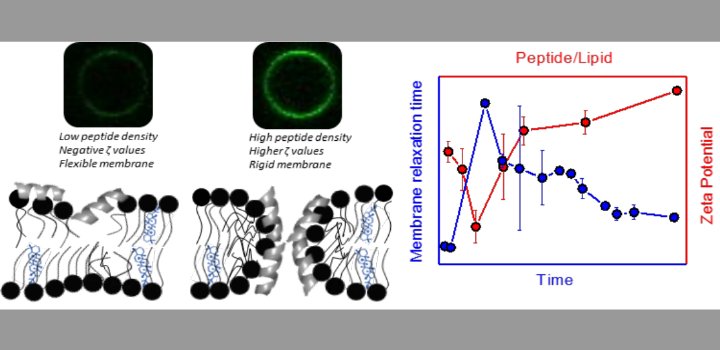
) for DP-containing LUVs showed a complex behavior at increasing peptide concentration. The effect of the peptide on membrane elasticity, investigated by nanotube retraction experiments, showed that peptide addition softened all membrane compositions, but membranes with DP got stiffer at long times. Considering this, and the ζ results, we propose that peptides accumulate at the interface adopting different arrangements, leading to a non-monotonic behavior. Possible correlations with cell membranes were inquired testing the antimicrobial activity of Polybia-MP1 against hopanoid-lacking bacteria pre-incubated with DP or CHO. The fraction of surviving cells was lower in cultures incubated with DP compared to those incubated with CHO. We propose that the higher activity of Polybia-MP1 against some bacteria compared to mammalian cells is not only related to membrane electrostatics, but also the composition of neutral lipids, particularly the hopanoids, could be important.
Keywords: Targeting of AMPs, Bacterial resistance, Membrane dynamic elasticity, Optical tweezers, Lytic activity, Zeta potential
Authors:
Article:
Alvares D.S, Monti M.R., Ruggiero Neto J., N Wilke. BBA Advances Volume 1, 2021, 100002. https://doi.org/10.1016/j.bbadva.2021.100002
There are several examples of single mutations that lead to a well-defined disease through a well-known mechanism. In other cases, a collection of mutations of the same protein produces a pathology with different degrees of severity. The accompanying work by Uusitalo et al. studies several mutants of the fatty acid binding protein P2 of the peripheral nervous system myelin. They conserve the native tertiary structure but a remarkable difference in the capacity to interact with lipids. This could be a clue to unravel the complex way in which these mutations affect myelin structure and function in a variant of Charcot-Marie-Tooth disease. Comment on: https://doi.org/10.1111/febs.16079.
Keywords: FABP; P2; emergence; myelin disorders; protein-lipid interaction.
Authors:
Article:
Pusterla J, Montich GG, Oliveira RG. Unraveling the etiology of myelin disorders: the P2 case in Charcot-Marie-Tooth disease. FEBS J. 2021 Jul 29. doi: 10.1111/febs.16131. Epub ahead of print. PMID: 34327848.

Zalosnik et al. Sci Rep. 2021

Nigra et al. Oxid Med Cell Longev. 2021

Alvarez Bolaño et al. Colloids SurfB 2021
Alvares et al. BBA Advances 2021

Nocelli NE et al. helion. 2021

Lardone and at. JBC 2021

Wagner et al. FASEB J. 2021
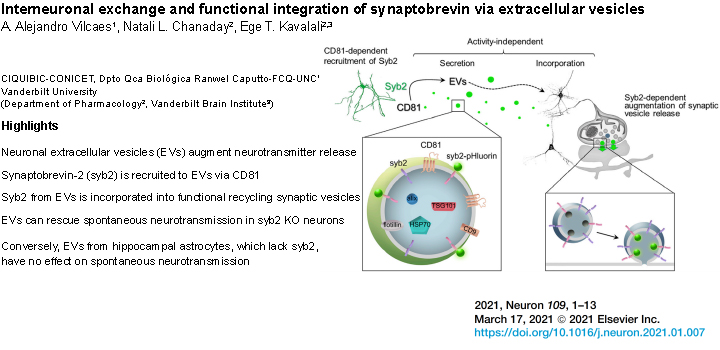
Vilcaes et al. J. Neuron. 2021

Cecchini NM et al. Plant J. 2020

Guido ME et al. 2020. Cell Mol Neurobiol

Prucca CG et al. Biochem J. 2020

Andrea G Albarracín Orio et al. Commun Biol. 2020

Zulueta Díaz AND BBA-Biomem. 2020
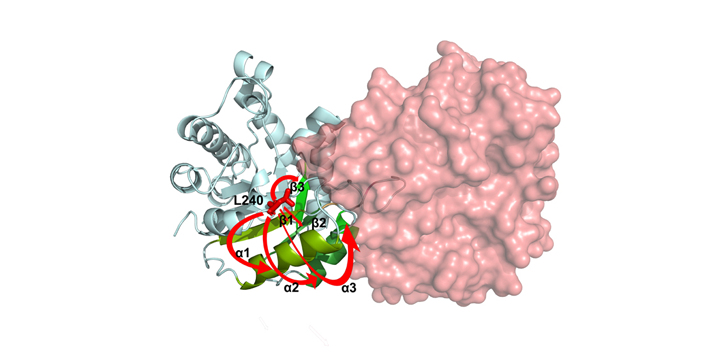
Romero J. Arch Biochem Biophys. 2020
Smith et al. Plant Physiol Biochem. 2020

Rodríguez-Berdini et al. J Biol Chem. 2020
Colque CA et al. Antimicrob Agents Chemother. 2020

Vilcaes et al. Int J Mol Sci. 2020

Cardozo Gizzi et al. Nat Protoc. 2020
Simultaneous observation of 3D chromatin organization and transcription at the single-cell level and with high spatial resolution may hold the key to unveiling the mechanisms regulating embryonic development, cell differentiation and even disease. We recently developed Hi-M, a technology that enables the sequential labeling, 3D imaging and localization of multiple genomic DNA loci, together with RNA expression, in single cells within whole, intact Drosophila embryos. Importantly, Hi-M enables simultaneous detection of RNA expression and chromosome organization without requiring sample unmounting and primary probe rehybridization. Here, we provide a step-by-step protocol describing the design of probes, the preparation of samples, the stable immobilization of embryos in microfluidic chambers, and the complete procedure for image acquisition. The combined RNA/DNA fluorescence in situ hybridization procedure takes 4–5 d, including embryo collection. In addition, we describe image analysis software to segment nuclei, detect genomic spots, correct for drift and produce Hi-M matrices. A typical Hi-M experiment takes 1–2 d to complete all rounds of labeling and imaging and 4 additional days for image analysis. This technology can be easily expanded to investigate cell differentiation in cultured cells or organization of chromatin within complex tissues.
Authors: Cardozo Gizzi AM, Espinola SM, Gurgo J, Houbron C, JB plug, DI Cats, Nollmann M.

Ruggiero et al. Immunol Cell Biol. 2020
Cardozo Gizzi A. et al. J Mol Biol. 2020

Chumpen S. et al. Biosci Rep. 2020
Malcolm M. et al. Front Cell Neurosci. 2019
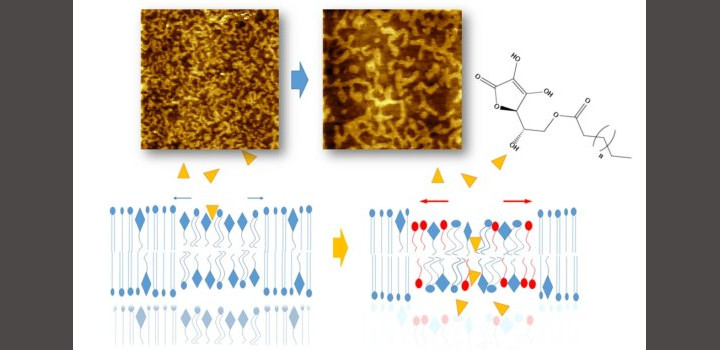
Zulueta Y. et al. Colloids Surf B Biointerfaces. 2020

Curtins JA et al. Biochem J. 2019
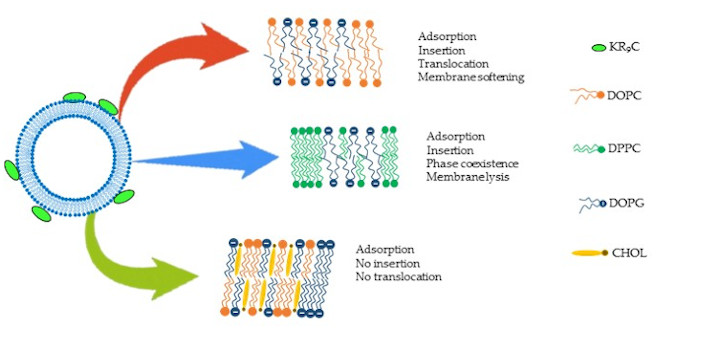
Crossio M. et al. Biomolecules. 2019
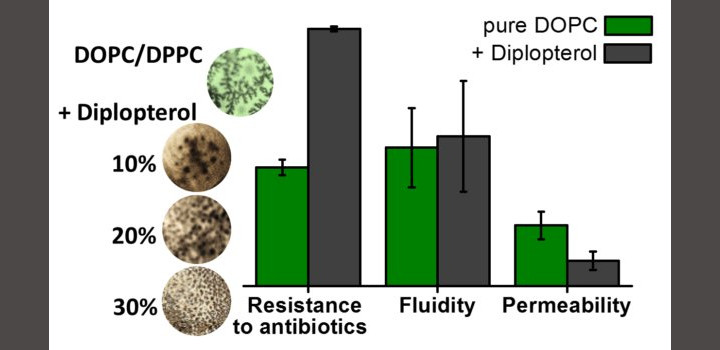
A Mangiarotti. et al. BBA-Biomembr. 2019
Rios M. et al. Front Cell Neurosci. 2019
Bertoldi ML et al. Front Cell Neurosci. 2019
Quiroga & Valdez Meth. Mol. Biol. 2019
Racca et al. Front. Oncol. 2019

Zlocowski et al. Sci. Rep. 2019

Cecchini et al. bio-protocol 2019
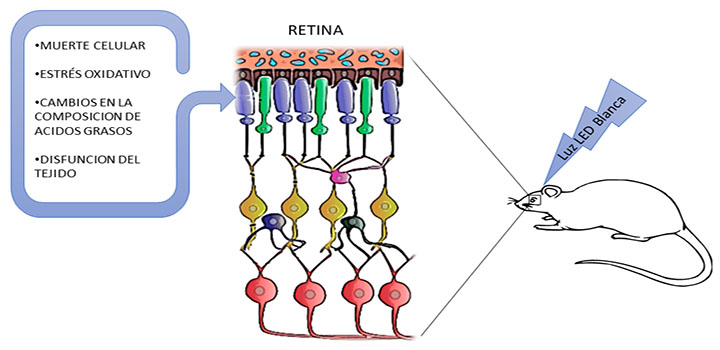
Benedetto, Front Cell Neurosci. 2019
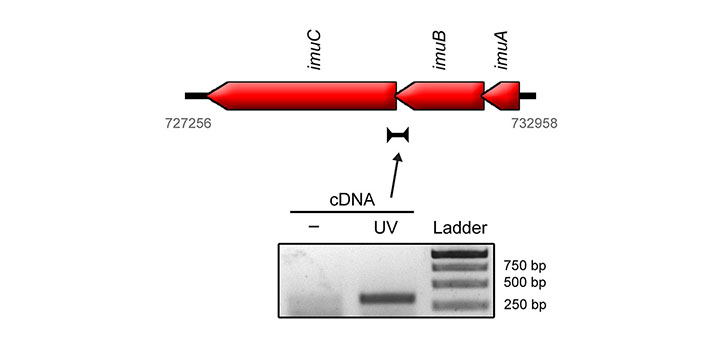
Luján et al. EnvirMolMut 2019

Szymanowski et al. BBA 2019

Castellaro et al. Cancers 2019
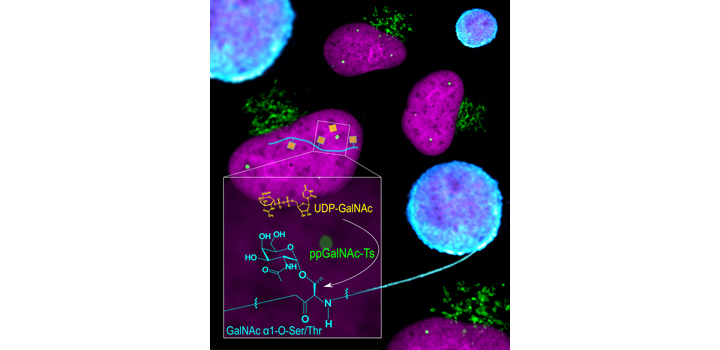
Cejas et al. JBC 2018

Deccaet to. BBA-Proteins 2018

Carpio & Katz. Methods Mol. Biol. 2019