Rizzi YS et al. 2016, Mol. Plant. Pathol.
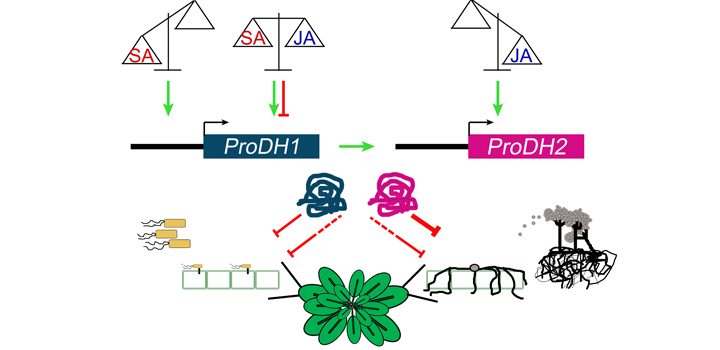
Arabidopsis contains two proline dehydrogenase (ProDH) genes, ProDH1 and ProDH2, encoding for homologous and functional isoenzymes. While ProDH1 has been extensively studied, especially under abiotic stress, ProDH2 began to be analyzed in recent years. These genes display distinctive expression patterns and show weak transcriptional co-regulation but they are both activated in pathogen infected tissues. We previously demonstrated that Arabidopsis plants silencing ProDH1/2 expression fail to trigger defenses against the hemibiotrophic bacterial pathogen Pseudomonas syringae pv. tomato AvrRpm1 (Pst-AvrRpm1), and showed that ProDH1 and ProDH2 have differential regulation by salicylic acid (SA). In the current work, we used prodh1 and prodh2 single mutant plants to assess the particular contribution of each gene to resistance against Pst-AvrRpm1 and the necrotrophic fungal pathogen Botyrtis cinerea (B. cinerea). In addition, we studied the sensitivity of ProDH1 and ProDH2 to the jasmonic acid (JA) defense pathway. We found that ProDH1 and ProDH2 are both necessary to achieve maximum resistance against Pst-AvrRpm1 and B. cinerea. However, ProDH2 has a major effect on early restriction of B. cinerea growth. Interestingly, ProDH1 is up-regulated by SA and JA, whereas ProDH2 is only activated by JA, and both genes display transcriptional inter-regulation at basal and infection conditions. These studies provide the first evidence on the contribution of ProDH2 to disease resistance, and describe a differential regulation and non-redundant but complementary functions of both enzyme isoforms in infected tissues, providing support for a fundamental role of ProDH in the control of biotrophic and necrotrophic pathogens.
Autores: Rizzi YS, Cecchini NM, Fabro G, Alvarez ME.
Artículo: Rizzi YS et al, Mol Plant Pathol. 2016 Aug 16. doi: 10.1111/mpp.12470.